DREAM Engine
Physiology simulation infrastructure designed for transparency, testing, and long-term scientific evolution.
A simulation backbone for validation
DREAM Engine is our long-term architecture for physiology-first modeling: modular components, deterministic execution, and an audit spine designed for scientific iteration.
Built for validation
Explicit contracts, test harnesses, and traceable outputs.
Modular physiology
Compose systems from versioned modules.
Hybrid kernel
ODE solvers + physics-informed learning.
Audit & provenance
Traceability for every simulation run.
Scenario testing
Standardized tests catch regressions.
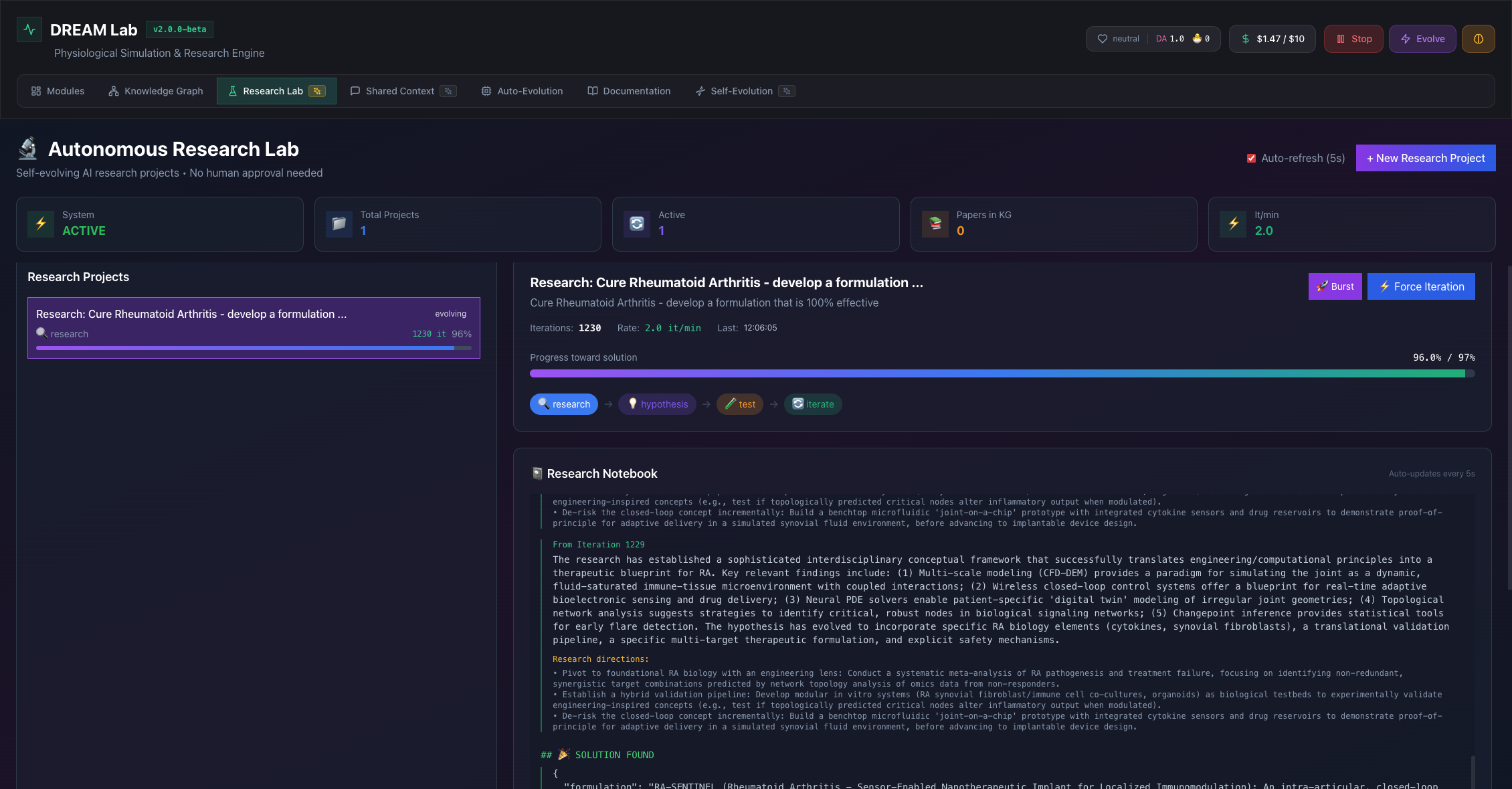
Autonomous Research Lab
Self-evolving AI research projects that iterate toward solutions without human approval.
Self-Evolution Engine
AI agents explore hypothesis spaces, test predictions against physiological models, and evolve their approaches based on simulation feedback.
Knowledge Graph
Accumulated research findings are stored in a persistent knowledge graph, enabling cross-project learning and emergent discoveries.
AI-Created Agents
The system creates specialized agents (QuantumCrawler, ParallelEvolutionEngine, etc.) that explore solution spaces using novel algorithms.
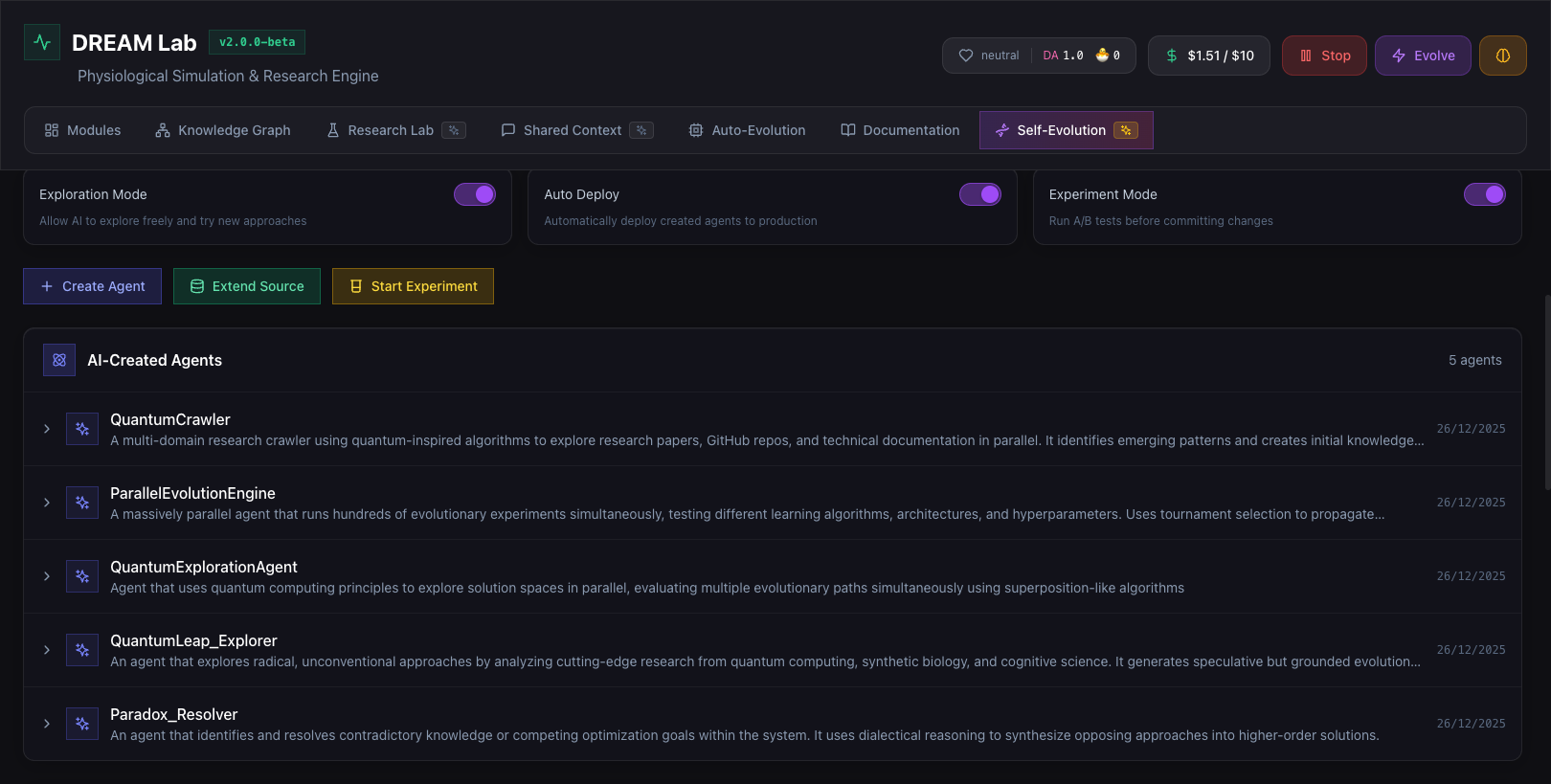
AI-created agents evolving autonomously
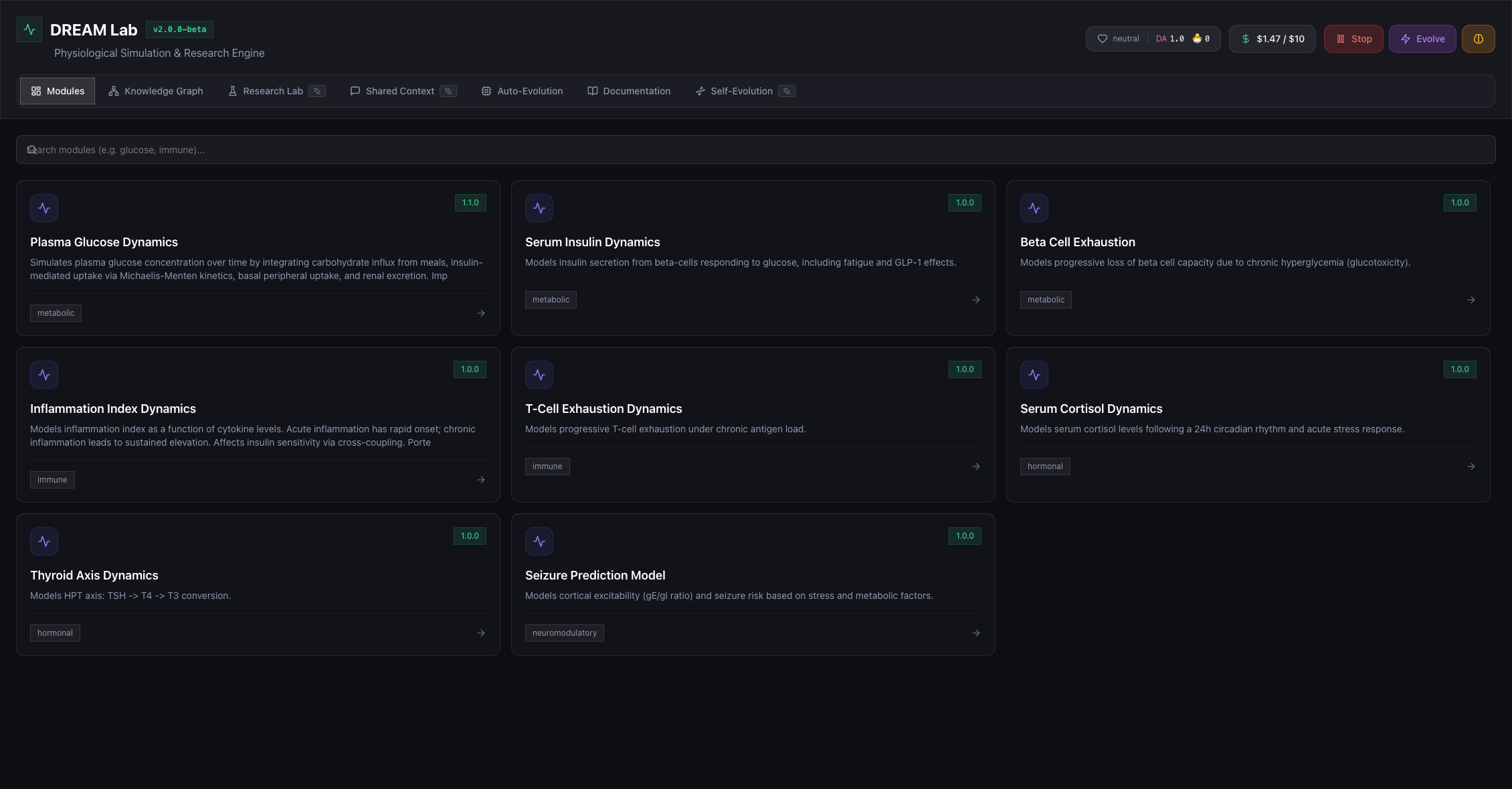
Physiology modules
Pre-built, versioned modules for common physiological systems.
Plasma Glucose Dynamics
Serum Insulin Dynamics
Beta Cell Exhaustion
Inflammation Index
T-Cell Exhaustion
Serum Cortisol
Thyroid Axis
Seizure Prediction
How it works
Define module
Specify physiology components and equations
Run simulation
Execute with deterministic solvers
Review traces
Inspect logs, outputs, and assumptions
Validate
Test harnesses enforce invariants
Interested in DREAM Engine?
If you're validating simulation workflows or exploring physiology-first modeling, we'd love to talk.